Recently published in the BMC Bioinformatics Journal, the research by Ranjan Kumar Barman, Anirban Mukhopadhyay, Ujjwal Maulik, and Santasabuj Das sheds light on the persisting challenge of infectious diseases. Despite significant progress in healthcare, these ailments, caused by a variety of pathogens, continue to burden public health and economies, particularly in low and middle-income countries. Understanding the intricate interaction between pathogens and host cells is pivotal in unraveling the complexities underlying infectious diseases.
Exploring Predictive Approaches for Disease-Associated Genes
Current genetic studies have predominantly revolved around single nucleotide polymorphisms (SNPs) and the identification of disease-associated genes available in public databases. However, the bulk of these efforts has concentrated on Mendelian diseases or complex conditions like asthma, diabetes, and cancer, leaving a notable gap in predicting host genes specifically associated with infectious diseases.
Leveraging Machine Learning in Gene Prediction
Addressing this gap, the study pioneers a new approach utilizing machine learning techniques, notably Deep Neural Networks (DNN). These techniques, recognized for their efficacy in diverse problem-solving, were compared against traditional classifiers such as Support Vector Machine, Naïve Bayes, and Random Forest. The research validated the model’s performance using independent datasets and ventured into unexplored territory by applying the model to proteins yet unutilized.

The labyrinthine nature of pathogen-host interactions poses a challenge in understanding the mechanisms behind infectious diseases. However, computational approaches offer an efficient and cost-effective avenue to identify disease-associated genes, capitalizing on the wealth of information available in public repositories.
Unveiling the Complexities of Host-Pathogen Interactions
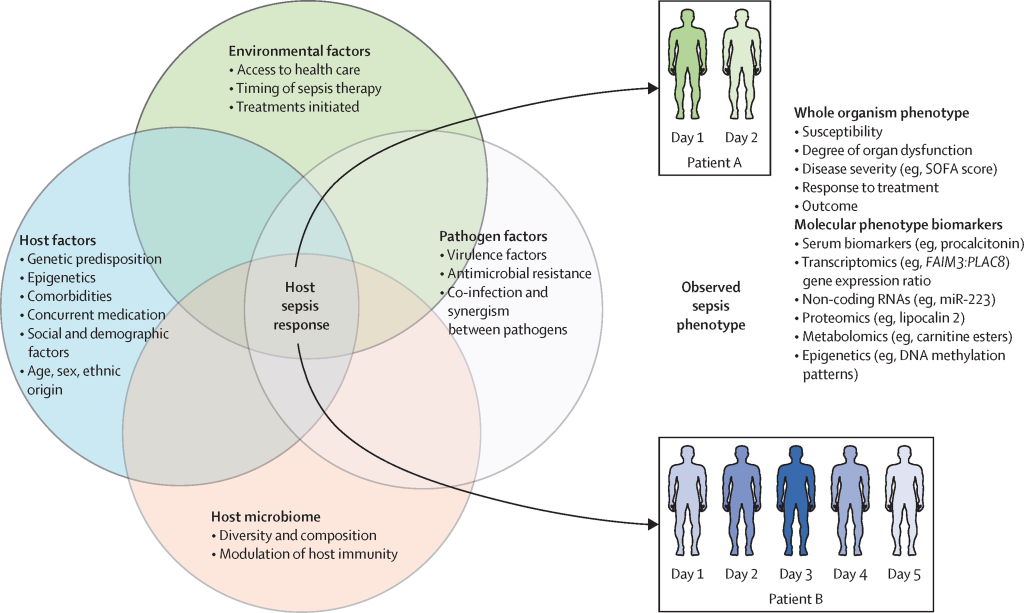
Despite the diverse clinical features of infectious diseases, they share commonalities like acute onset, transmissibility, unique immune responses, and varied reactions to antimicrobial agents. Host responses to infections significantly differ from non-infectious diseases, involving specific molecular patterns that trigger innate immune receptors. This study integrated sequence and protein-protein interaction (PPI) network properties, revealing the dominance of the latter in predicting infectious disease-related host genes.
The Ascendancy of Deep Learning
Deep Neural Networks, particularly through the use of TensorFlow, demonstrated promising results, achieving high accuracies during training. However, their performance slightly decreased during testing, emphasizing their suitability for larger datasets and features. Ensemble feature selection techniques bolstered predictive capabilities, performing on par with using all features.

Validating Comparative Analysis
The model’s validity was established through comparisons with existing machine learning techniques (MLT)-based methods designed for other diseases like cancer and Alzheimer’s. The infectious disease-associated gene prediction model outperformed these disease-focused models, affirming its reliability in identifying disease-associated host genes.

Insights and Future Implications
This study identified numerous infectious disease-associated host genes, laying the groundwork to expand our comprehension of disease pathogenesis. The discovered genes correlated with critical biological processes and disease ontology, suggesting potential common pathways among various diseases. This opens avenues for repurposing existing treatments to develop new host-targeted therapies for infectious diseases.
Conclusion
The proposed computational model, integrating sequence and PPI network properties, demonstrates its efficacy in predicting infectious disease-associated host genes. This method achieves high accuracy, emphasizing its potential in identifying disease risks and therapeutic targets, paving the way for a deeper understanding of diseases and innovative therapeutic strategies.
Study DOI: 10.1186/s12859-019-3317-0
Engr. Dex Marco Tiu Guibelondo, B.Sc. Pharm, R.Ph., B.Sc. CpE
Subscribe
to get our
LATEST NEWS
Related Posts

Bioinformatics & Multiomics
Harnessing Computational Ingenuity for Tomorrow’s Therapeutics
Leverage computational power to navigate modern drug design.
Bioinformatics & Multiomics
Exploring Cutting-Edge Techniques in Protein Target Identification
From genetic screens in model organisms to chemical proteomics in mammalian systems, each approach offers unique insights into the complex landscape of drug-target interactions.
Read More Articles
Synthetic Chemistry’s Potential in Deciphering Antimicrobial Peptides
The saga of antimicrobial peptides unfolds as a testament to scientific ingenuity and therapeutic resilience.