A new computational analysis method for profiling more than a million tandem repeats (TRs) throughout the human genome using PacBio‘s very own native long-read HiFi sequencing data became available in Q3 of 2022. PacBio (NASDAQ: PACB) is a leading provider of high-quality, highly accurate sequencing solutions. Scientists can now fully characterize the sequence and methylation status of tandem repeats (TRs) genome-wide thanks to the Tandem Repeat Genotyping Tool (TRGT; pronounced “target”). The use of TRGT is thought to help researchers better understand how known TRs contribute to human disease and may help identify new TRs that cause disease.
The Key to Understanding is Repetition
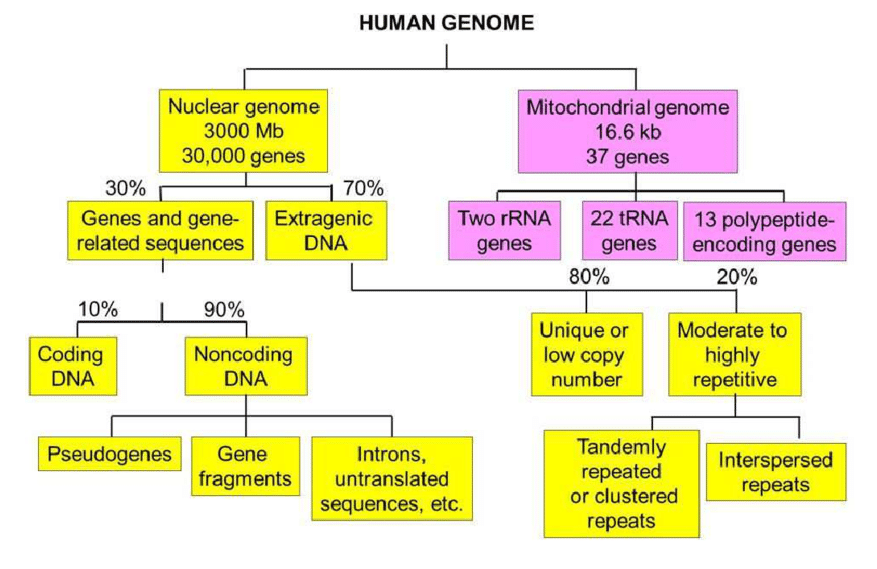
Typically, repetitive DNA is described as DNA that exists in the genome in several copies but has no known biological purpose. Tandem repetitions (tandem repeats) and scattered repeats (interspersed repeats) are the two basic categories of repeated elements.
Interspersed repeats, which make up about 45% of the human DNA, are the remains of transposable elements (TEs). TEs are either nonautonomous, amplified by proteins encoded by nonautonomous elements, or autonomous, encoding proteins required for their amplification. The majority of interspersed repetitions are severely altered, old TEs that cannot transpose. A few families of non-LTR retrotransposons, such as the Alu and LINE1 (L1) elements, continue to actively replicate in the human genome and sporadically insert into human genes.
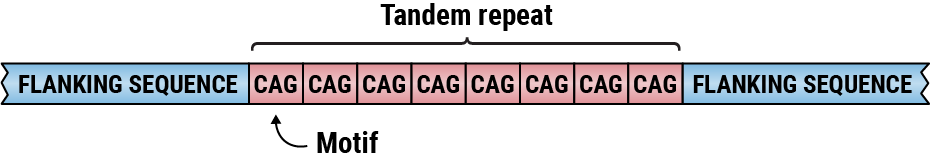
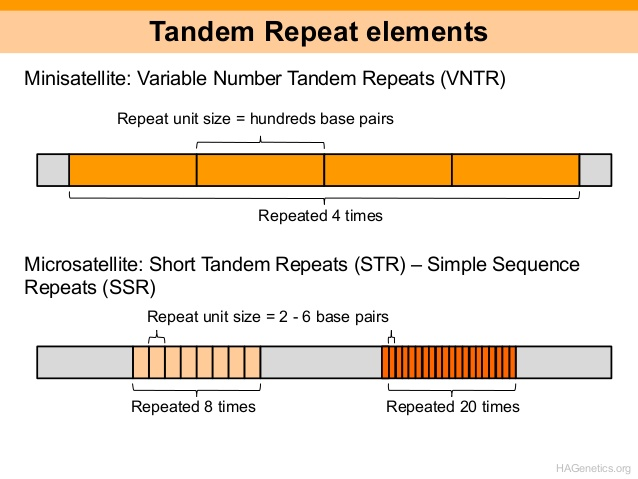
Tandem repeats are head-to-tail arrangements of the same sequence motifs repeated repeatedly. Tandem repeats are simply repeating nucleotide sequences where the copies are close to one another. One or more nucleotides could be repeated. For instance, the tandem repeat CG CG CG CG CG repeats the sequence CG five times.
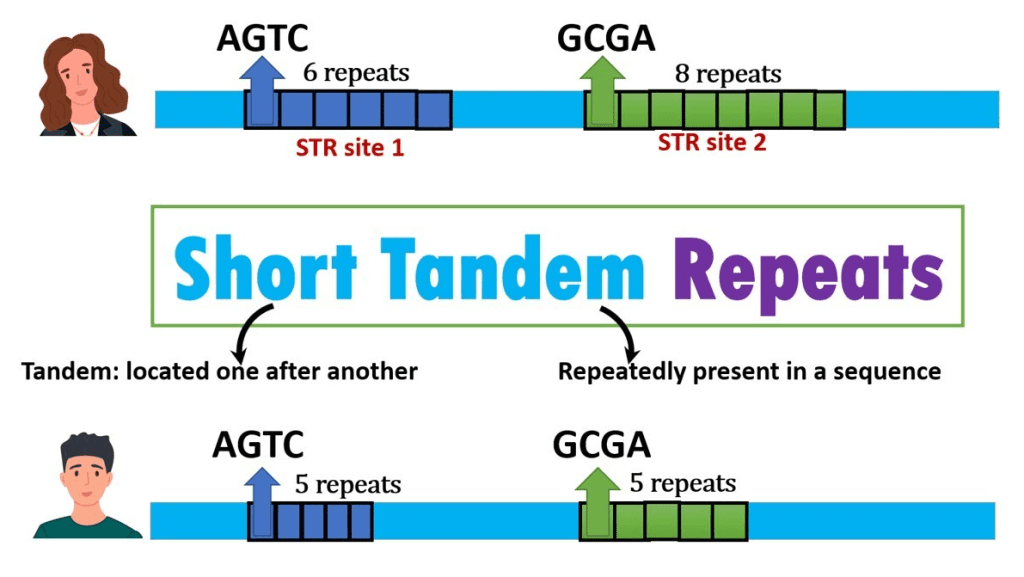
Satellite DNAs, microsatellites, and minisatellites all contain tandem repeats. Satellite DNA is a tandem repeat with mostly non-coding DNA copies. It is primarily found in heterochromatin and centromeres. A microsatellite is a short tandem repeat with a length of 1-6 base pairs or more, which is frequently repeated five to fifty times. A minisatellite is similar to a tiny tandem repeat in that it is repeated five to fifty times, but with considerably longer base pairs—from 10 to 60. The roughly 500-copy 40S rRNA gene found in toad somatic cells is an example of a tandem repeat.
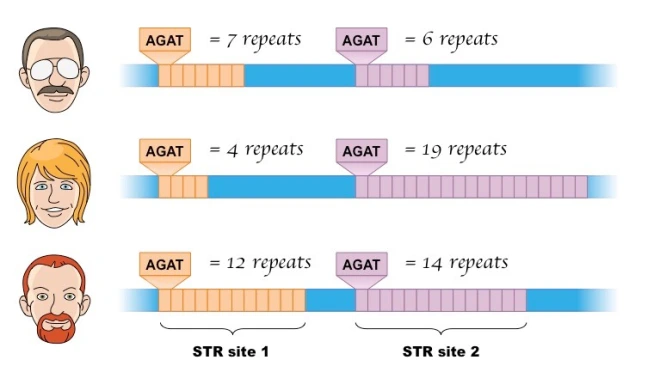
Tandem repeats can occasionally be used as genetic markers to trace family ancestry. From parent to child, they might increase in size.
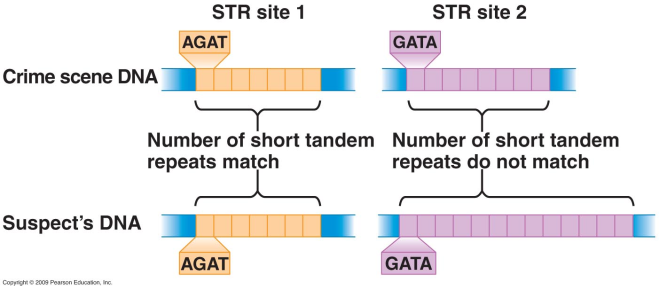
In forensic studies, they can also be helpful for DNA fingerprinting. Importantly, TRs have been connected to numerous neurological conditions, including ALS and Huntington’s disease, in addition to Fragile X syndrome, the leading cause of inherited mental intellectual disability.
Improved Characterization for Better Comprehension
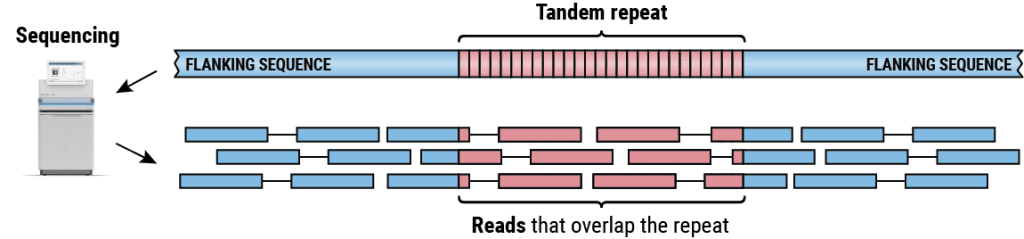
Tandem repeats are one of the most challenging variant classes to describe genetically and epigenetically, according to Michael Eberle, Vice President of Computational Biology at PacBio. Tandem repeats have not received enough attention up to this point because short-read sequencing methods cannot yet sequence these parts of the genome. The project aims to enable scientists to investigate and describe these complex genomic areas and, ultimately, better understand their biological significance by combining HiFi sequencing and TRGT.
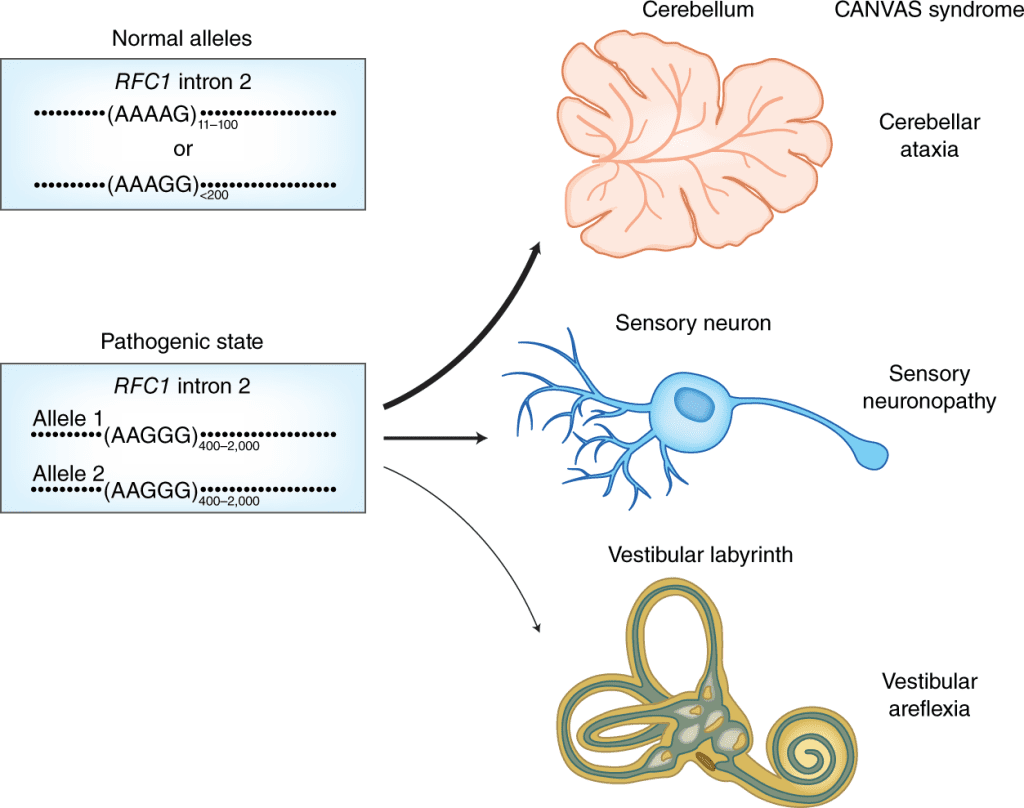
The goal of TRGT is to provide research scientists with the tools necessary to characterize the sequence’s composition and structure, repeat unit length, and CpG methylation for each analyzed repeat allele and flanking sequence throughout the genome. 5’—C—phosphate—G—3′ stands for the nucleosides cytosine and guanine, which are connected by just one phosphate group in DNA. In the tertiary analysis for loci causing diseases, the improved characterization of TR variation may be helpful. TRGT, for instance, can identify the extremely lengthy (thousands of base pair) repeats connected to particular disorders. TRGT can also detect alterations in sequence composition that may be linked to pathogenic expansions in conditions such as cerebellar ataxia with neuropathy and bilateral vestibular areflexia syndrome (CANVAS). TRGT can also recognize hypermethylation signals like those seen with Myotonic Dystrophy expansions since HiFi readings can detect CpG methylation.
In data from people with inherited illnesses, the TRGT method is a significant improvement over repeat expansion analysis and is a useful tool for finding novel and possibly significant variations that may be related with disease. Stephan Zuchner, MD, PhD, FAAN, Professor and Chief Genomics Officer at the Miller School of Medicine, University of Miami, makes this point quite clearly. Research into the characterization of repeat expansions in healthy and uncommon illness cohorts is the main area of interest for Dr. Zuchner and his colleague Matt Danzi, PhD, a computational neurobiologist.
TRGT also includes a companion tool, TRVZ, that enhances usability by displaying read pileups and methylation data for each repeat allele and flanking sequence studied. Now accessible on GitHub at https://github.com/pacificBiosciences/trgt, TRGT and TRVZ
About PacBio
The American biotechnology company Pacific Biosciences of California, Inc. (also known as PacBio) was established in 2004 and specializes in the development and production of systems for real-time biological observation and new gene sequencing. Single-molecule real-time sequencing (SMRT), a technology developed by PacBio, is based on the characteristics of zero-mode waveguides.
The business was started as a result of research at Cornell University that merged studies in photonics, semiconductor processing, and biology. The initial workers were Steve Turner, Mathieu Foquet, and Jonas Korlach, three graduate students in Professors Watt W. Webb’s and Harold Craighead’s lab.
Nanofluidics, Inc. was the company’s original name.
Prior to its initial public offering in October of that year, the firm raised almost US$400,000,000 in six rounds of financing, largely from venture capital, making it one of the most capitalized startups in 2010. Mohr Davidow Ventures, Kleiner Perkins, formerly Kleiner Perkins Caufield & Byers (KPCB), Alloy Ventures, and Wellcome Trust were significant investors.
The PacBio RS, the company’s first commercial product, was offered to a select group of clients in 2010 before becoming on sale in early 2011. The PacBio RS II, an updated version of the sequencer, was made available in April 2013.
Currently, PacBio is regarded as a leading life science technology firm that designs, develops, and produces advanced sequencing technologies to aid researchers and clinicians in solving genetically complex issues. The PacBio technologies and products currently in development are based on two highly distinctive core technologies that are centered on precision, completeness, and quality. These technologies are PacBio’s HiFi long read sequencing, which is already in use, and SBBTM short read sequencing, which is currently under development. Infectious disease and microbiology, oncology, plant and animal sciences, human germline sequencing, infectious disease, and other new applications are only a few of the scientific problems that PacBio products address.
For more information, please visit www.pacb.com and follow @PacBio.
Products from PacBio are only available for research purposes. Use not intended for diagnostic procedures.
Subscribe
to get our
LATEST NEWS
Related Posts

Bioinformatics & Multiomics
Harnessing Computational Ingenuity for Tomorrow’s Therapeutics
Leverage computational power to navigate modern drug design.
Bioinformatics & Multiomics
Exploring Cutting-Edge Techniques in Protein Target Identification
From genetic screens in model organisms to chemical proteomics in mammalian systems, each approach offers unique insights into the complex landscape of drug-target interactions.