In vitro biological research has seen a significant shift towards the adoption of 3D cell culture models, including cellular aggregates, microtissues, spheroids, and complex organoid models. These 3D culture systems offer distinct advantages over traditional 2D cultures, such as better physiological cellular organization, enhanced cell-to-cell and cell-to-matrix communications, and improved cellular differentiation and signaling.
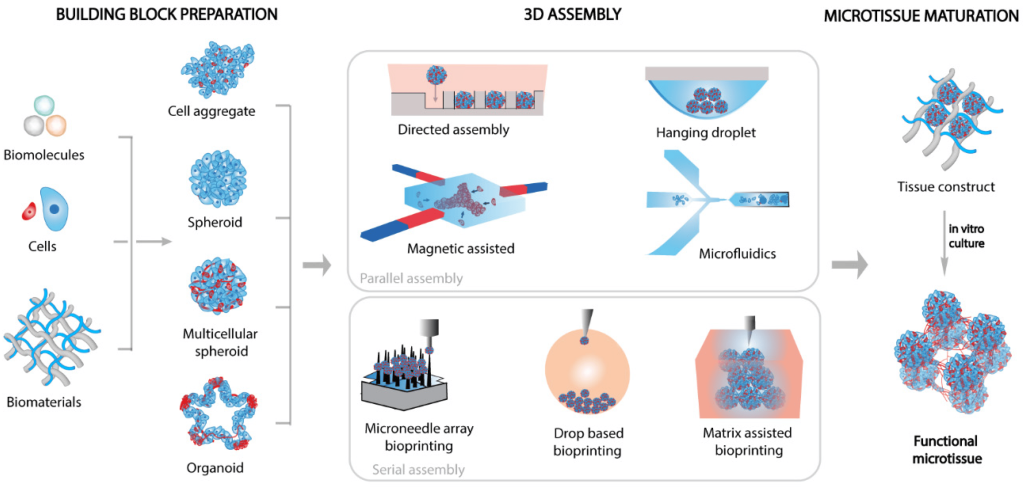
Organoids, in particular, are complex 3D systems that mimic tissue-specific organogenesis, cellular heterogeneity, cytoarchitectures, and functions. Consequently, 3D culture systems hold great promise for fundamental research, drug development, personalized medicine, and regenerative medicine.
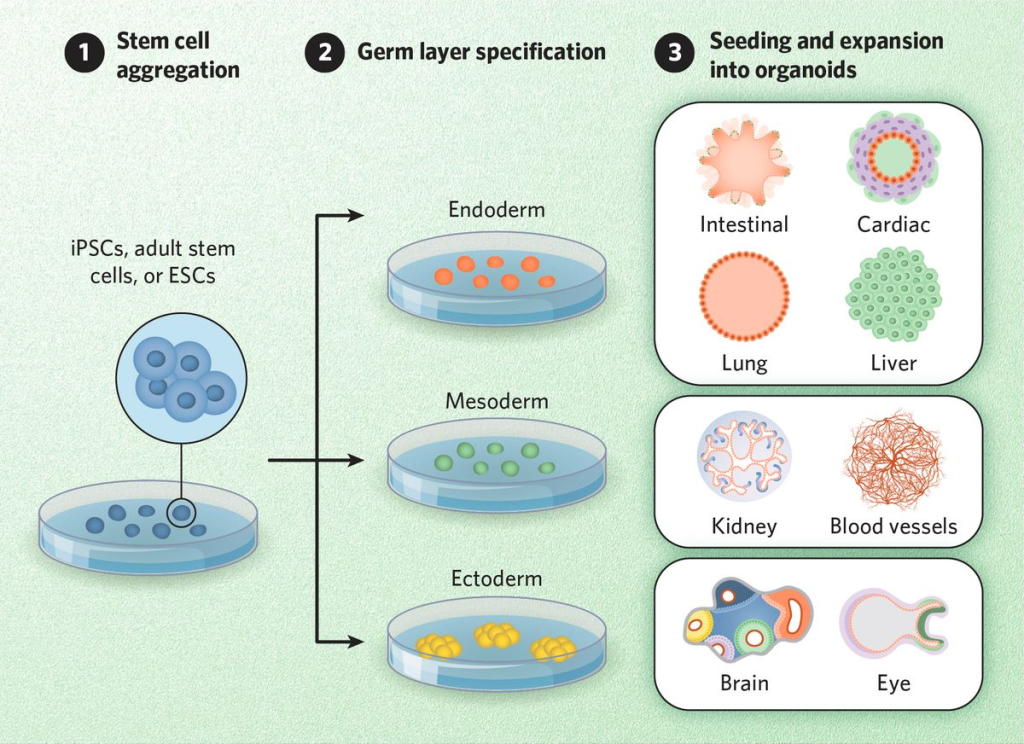
Advances in 3D Culture Methods
The growing popularity of 3D cultures has spurred the development of various methods and protocols for their generation. These methods include low-adherence culture plates for 3D cell culture in suspension, bioreactors that allow large-volume dynamic flow conditions to improve oxygenation, and microfluidic chips known as organ-on-chip systems that precisely control fluid flows and replicate the cellular microenvironment.
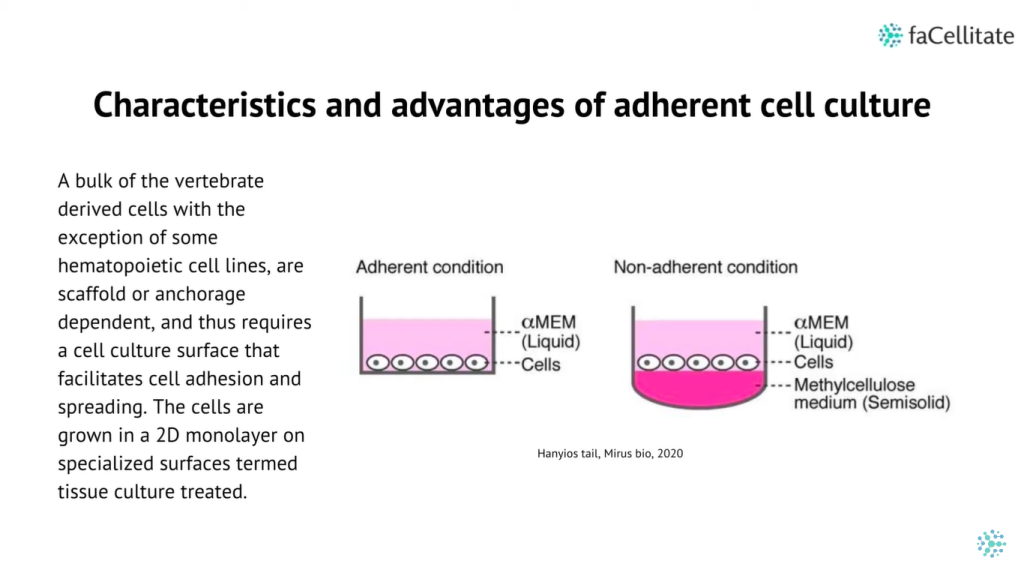
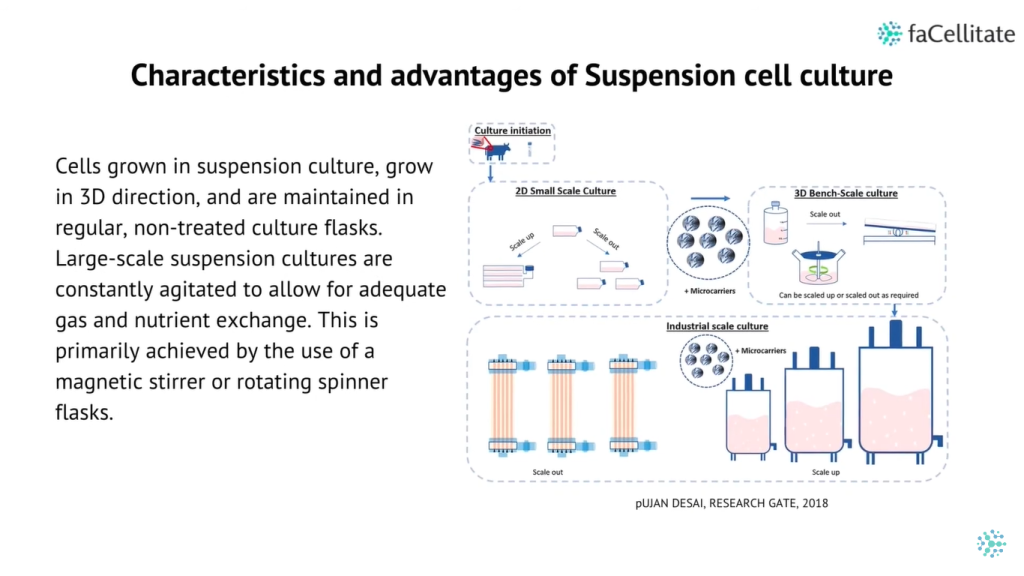
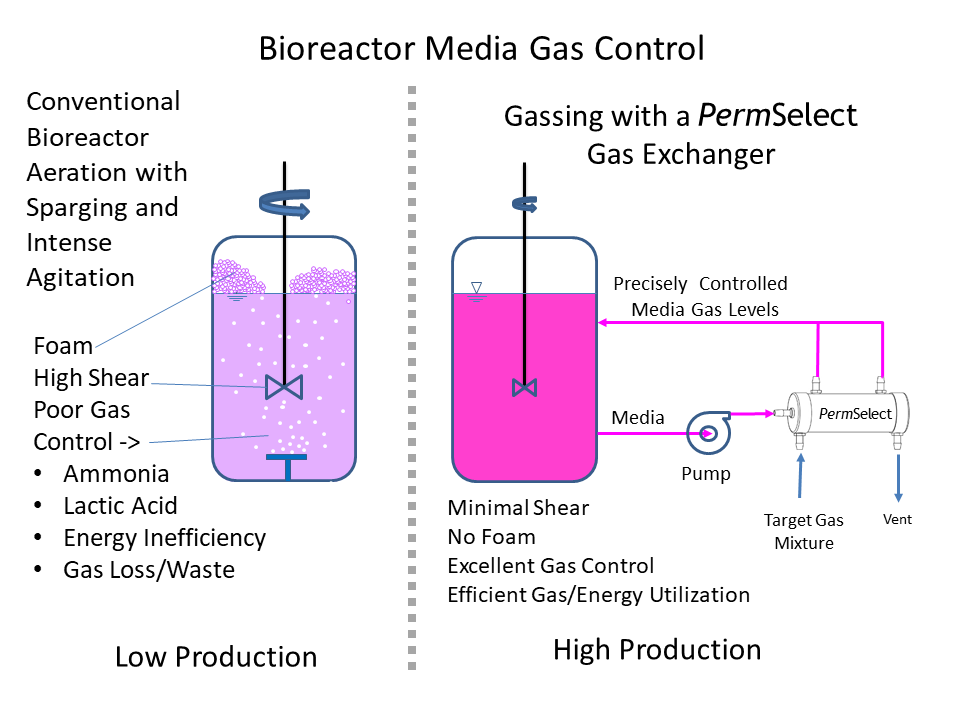
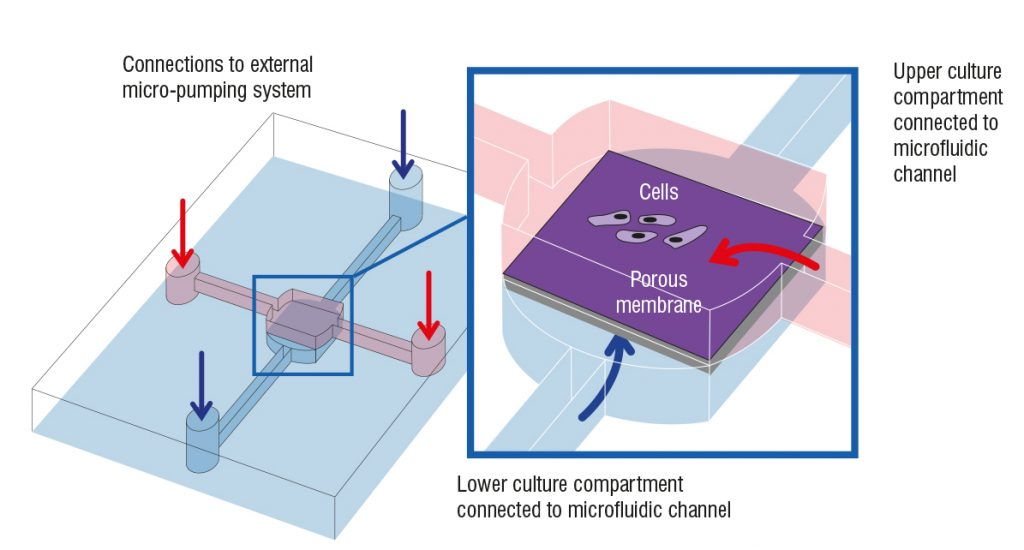
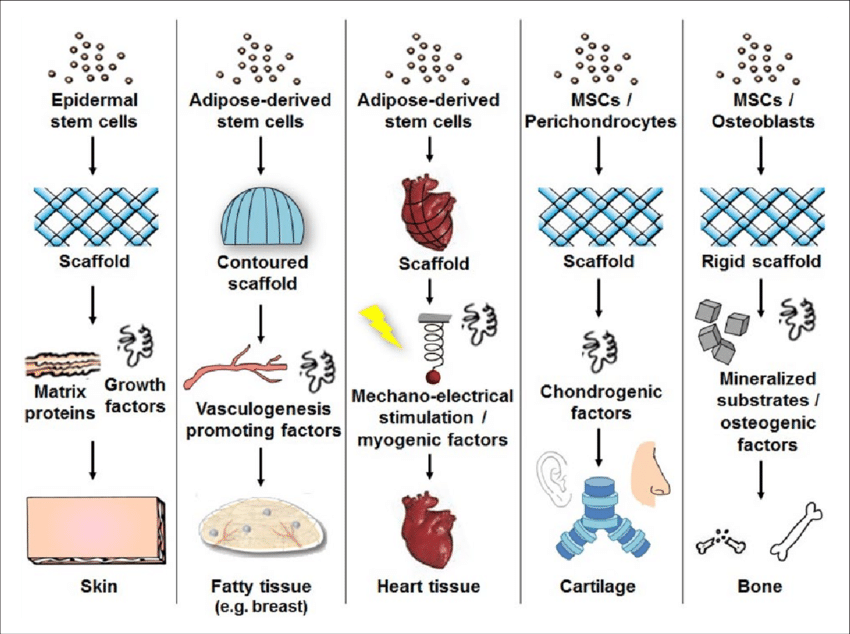
Additionally, bioengineering strategies such as the use of 3D scaffolds and bioprinting have provided researchers with greater control over critical parameters, including fluid flows, nutrient and oxygen supply, and waste management.
doi: 10.1177/1753193415571308.
Challenges in Transitioning from 2D to 3D Cultures
Transitioning from 2D to 3D culture systems presents unique challenges, particularly in assessing cell viability over time. Historically, lactate dehydrogenase (LDH) activity assays have been used to assess cellular viability in 2D cultures. LDH, a cytoplasmic enzyme present in almost all cell types, is released into the extracellular environment when the cell membrane is damaged. The LDH activity assay is based on the enzymatic conversion of lactate to pyruvate by LDH, resulting in the production of a detectable colored or luminescent molecule. The amount detected correlates with the cell viability in the culture conditions.
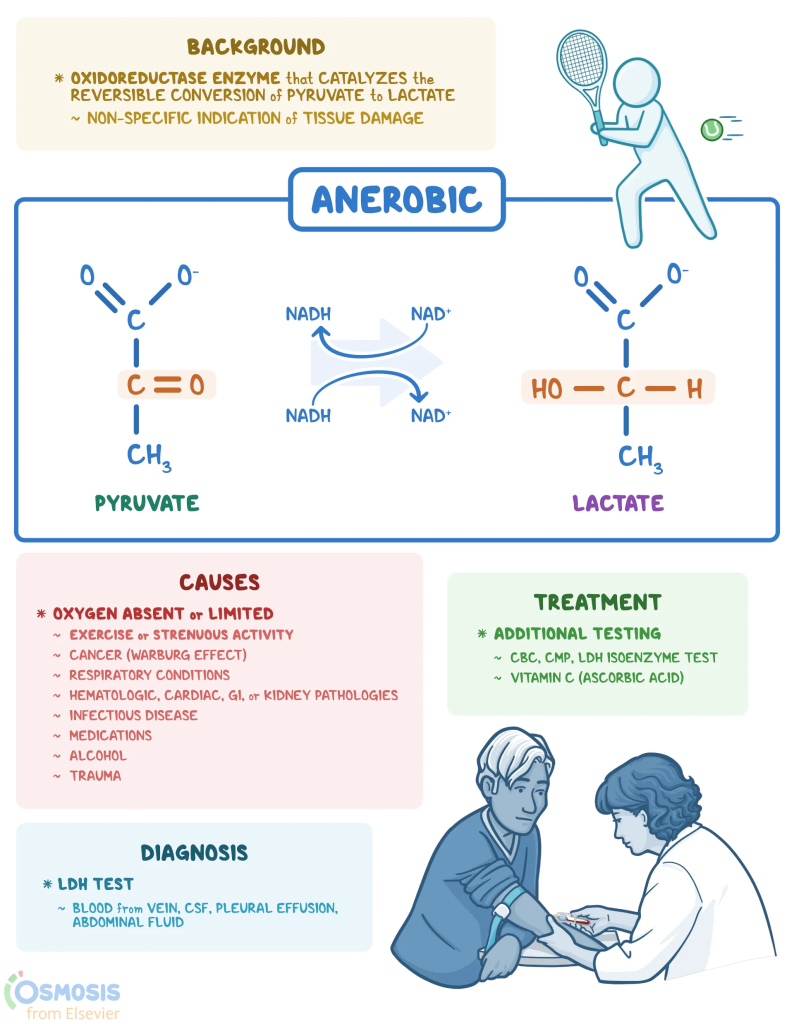
Implementing the LDH assay in 3D cell cultures faces several challenges, including the need for a robust normalization step. In 2D cultures, LDH activity is usually normalized by the number of seeded cells, the LDH activity in cell lysates, or the protein concentration. However, the complexity of 3D cultures makes precise quantification of cell density difficult while preserving culture integrity. Normalizing LDH activity by protein concentration in the conditioned culture medium has emerged as a potential strategy.
Stabilizing LDH Activity for Longitudinal Studies
LDH activity is not stable over time, requiring quantification on fresh conditioned medium for accurate results. This poses a substantial obstacle to longitudinal comparisons. Using a storage buffer to preserve LDH activity at temperatures below −20 °C for several days is one solution. To address these limitations, an optimized LDH activity assay protocol specifically tailored for 3D cell cultures has been developed. This approach integrates a normalization method based on the quantification of total protein content in the conditioned culture medium and the characterization of a dedicated preservation buffer capable of maintaining LDH stability at −20 °C for one month.
Methodology for LDH Assay Optimization
Human-induced pluripotent stem cells (hiPSCs) were used to generate cerebral organoids following established protocols. The organoids were exposed to valproic acid, a known neurotoxin, at two concentrations to demonstrate the effectiveness of the optimized LDH assay in normalizing cytotoxicity levels between the organoids.
To preserve LDH activity, a conditioned medium (CM) was mixed with an LDH-preservation buffer containing Bovine Serum Albumin (BSA) as a stabilizing agent. Aliquots of CM sampled from cerebral organoids were frozen with or without the preservation buffer and analyzed monthly over four months. The preservation buffer comprised Tris-HCl buffer solution, glycerol, and BSA.
Results and Discussion
The optimized LDH assay demonstrated that the preservation buffer extended LDH stability for up to a month at −20 °C. Consistent LDH levels were observed prior to a decline after one month of storage compared to samples without the buffer. Loss of LDH activity in CM without the buffer was evident after one week at −20 °C, with a total loss of activity after three months.
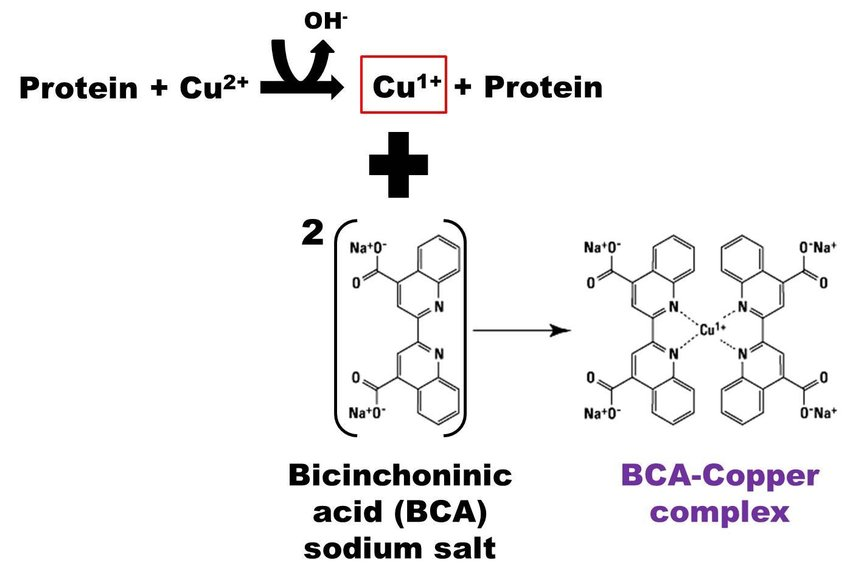
The presence of BSA in the preservation buffer necessitated adapted dilutions to prevent signal saturation. Bicinchoninic assay (BCA) with CMB samples diluted in distilled water indicated that the preservation buffer maintained the proportionality between protein concentration and absorbance, ensuring linearity. Discrepancies emerged when converting absorbance to protein concentration using a BSA standard curve. Luminescence-based LDH detection revealed signal saturation at the most concentrated dilutions. Optimal CMB dilutions ranged from 1/2.5 to 1/4 with distilled water.
The optimized LDH assay was validated with cerebral organoids exposed to valproic acid. Exposure to 10 mM VPA significantly elevated LDH levels compared to controls, indicating high cytotoxicity. Exposure to 1 mM VPA showed no significant impact on LDH levels, consistent with literature findings that VPA at this concentration impairs cortical development without affecting organoid morphological integrity.
A Standardized Framework for 3D Culture Analyses
The development of this refined LDH assay represents an advancement in addressing the challenges associated with longitudinal monitoring of 3D culture viability. By tailoring the standard LDH assay to accommodate the specific constraints of 3D systems, the proposed normalization and long-term storage methodologies facilitate the transition of this routine test from 2D to 3D. This enhanced LDH assay not only paves the way for the development of quantification techniques adapted to the analysis of conditioned media from 3D cultures, but also broadens the scope of cellular viability assessment in 3D. Integration of this method in routine characterization of 3D cultures holds great promise for enhancing the versatility of these models and for advancing various biomedical and toxicological applications that rely on cytotoxicity evaluations.
Engr. Dex Marco Tiu Guibelondo, B.Sc. Pharm, R.Ph., B.Sc. CpE
Subscribe
to get our
LATEST NEWS
Related Posts

Drug Discovery Biology
Proteolytic Rewriting: Engineering Controlled Absence of Pathogenic Protein Persistence
Targeted protein degradation transforms drug therapy by engineering the cellular machinery to erase, rather than merely inhibit, pathogenic proteins.

Drug Discovery Biology
Open Nucleotides: Grant-Driven Infrastructure for Equitable mRNA Vaccine Manufacturing
By developing accessible cap analogs and RNA raw materials, Hongene Biotech, guided by David Butler’s expertise in nucleotide chemistry and supported by the Gates Foundation, is reshaping the molecular infrastructure that underpins global mRNA vaccine equity.
Drug Discovery Biology
Redox Messengers: How Endogenous Gasotransmitters Rewire Vascular Biology to Resist Age-Driven Oxidative Stress
Gasotransmitters provide a biologically sophisticated means of counteracting age-related oxidative stress and preserving vascular resilience.
Drug Discovery Biology
Aptamer Signal Dynamics: Engineering Nucleic Acid Recognition Systems for High-Fidelity, Multi-Modal Biosensing
Aptamers redefine biosensing by pairing programmable molecular recognition with versatile transduction strategies capable of detecting clinically relevant biomarkers with exceptional fidelity.
Read More Articles
Spatial Collapse: Pharmacologic Degradation of PDEδ to Disrupt Oncogenic KRAS Membrane Localization
PDEδ degradation disrupts KRAS membrane localization to collapse oncogenic signaling through spatial pharmacology rather than direct enzymatic inhibition.
Neumedics’ Integrated Innovation Model: Dr. Mark Nelson on Translating Drug Discovery into API Synthesis
Dr. Mark Nelson of Neumedics outlines how integrating medicinal chemistry with scalable API synthesis from the earliest design stages defines the next evolution of pharmaceutical development.
Zentalis Pharmaceuticals’ Clinical Strategy Architecture: Dr. Stalder on Data Foresight and Oncology Execution
Dr. Joseph Stalder of Zentalis Pharmaceuticals examines how predictive data integration and disciplined program governance are redefining the future of late-stage oncology development.
Exelixis Clinical Bioanalysis Leadership, Translational DMPK Craft, and the Kirkovsky Playbook
Senior Director Dr. Leo Kirkovsky brings a rare cross-modality perspective—spanning physical organic chemistry, clinical assay leadership, and ADC bioanalysis—to show how ADME mastery becomes the decision engine that turns complex drug systems into scalable oncology development programs.
Policy Ignition: How Institutional Experiments Become Durable Global Evidence for Pharmaceutical Access
Global pharmaceutical access improves when IP, payment, and real-world evidence systems are engineered as interoperable feedback loops rather than isolated reforms.
Sepsis Shadow: Machine-Learning Risk Mapping for Stroke Patients with Bloodstream Infection
Regularized models like LASSO can identify an interpretable risk signature for stroke patients with bloodstream infection, enabling targeted, physiology-aligned clinical management.