A groundbreaking paradigm has emerged In the ever-evolving landscape of biomedical research, promising to unravel the intricacies of biological systems at a level never before possible. The approach, known as multi-omics, transcends the limitations of traditional single-omics studies by integrating data from genomics, transcriptomics, proteomics, metabolomics, and epigenomics. This comprehensive method, though laden with promises, encounters formidable challenges in data heterogeneity, integration complexities, and the need for advanced computational methods. This article delves into the recent strides and hurdles of multi-omics, focusing on its applications in shaping the future of pharmaceutical sciences and innovative therapeutic development.
The Pinnacle of Integration: Challenges in Multi-Omics Approaches
The journey of multi-omics begins with the collection of data from various biological levels, samples, conditions, and technologies. However, the richness of this data brings forth challenges in integration, analysis, and interpretation. The high dimensionality and complexity demand sophisticated computational tools and statistical methods, posing a formidable barrier for researchers. The biological interpretation and validation of multi-omics results further necessitate profound knowledge and experimental verification in the field of interest.
Navigating Complexity: Approaches for Multi-Omics Data Integration
To surmount the challenges, researchers employ diverse integration strategies, each tailored to the nature of the data and the underlying biological questions. Four predominant approaches have gained prominence: conceptual integration, statistical integration, model-based integration, and networks and pathway data integration. These methodologies serve as the scaffolding for unraveling the complexity of multi-omics data, allowing researchers to bridge the gap between disparate datasets and glean meaningful insights into biological systems.
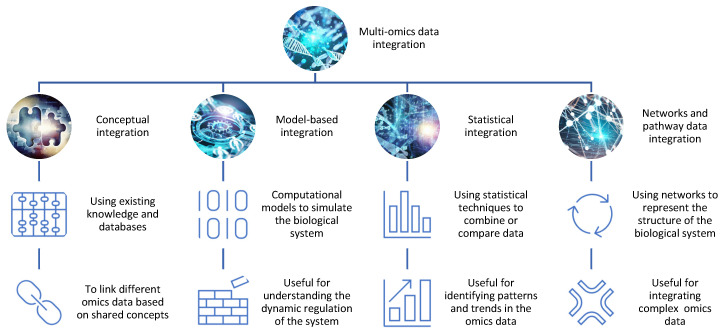
Conceptual Integration: Bridging Through Shared Concepts
Conceptual integration relies on leveraging existing knowledge and databases to link omics data based on shared concepts such as genes, proteins, pathways, or diseases. Researchers use tools like STATegra and OmicsON to enhance the framework’s capacity to detect specific features overlapping between compared omics sets. While powerful for generating hypotheses and exploring associations, it may fall short in capturing the dynamic nature of biological systems.
Statistical Integration: Unveiling Patterns Through Quantitative Measures
Statistical integration harnesses quantitative measures like correlation, regression, clustering, or classification to combine or compare different omics data. This approach unveils patterns and trends in the data but may struggle to reveal causal or mechanistic relationships between the integrated datasets.
Model-Based Integration: Simulating Biological Dynamics
Model-based integration employs mathematical or computational models to simulate or predict biological system behavior based on different omics data. While insightful for understanding system dynamics and regulation, it demands extensive prior knowledge and assumptions about system parameters and structure.
Networks and Pathway Data Integration: Charting Complexity Through Networks
Networks and pathway data integration deploy graphical representations to illustrate the structure and function of biological systems. Using protein–protein interaction (PPI) networks or metabolic pathways, this method excels in integrating multiple omics data types at varying levels of granularity and complexity, though temporal and spatial aspects may be overlooked.
Decoding the Molecular Symphony: Applications of Multi-Omics in Drug Discovery
Multi-omics stands at the forefront of revolutionizing drug discovery, promising unparalleled insights into molecular mechanisms and interactions. The ability to identify and validate drug targets and biomarkers represents a pivotal application, and multi-omics achieves this through various means.
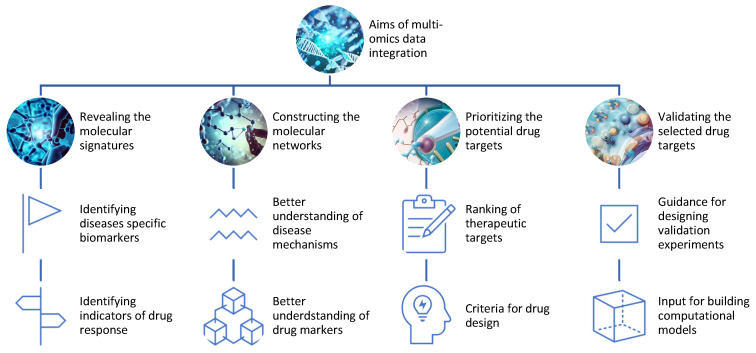
Revealing Molecular Signatures: Unraveling Disease and Drug Responses
Multi-omics unveils the molecular signatures of diseases and drug responses by identifying differentially expressed genes, proteins, metabolites, and epigenetic marks. Through this, researchers gain a comprehensive understanding of the molecular landscape, differentiating between healthy and diseased states or responsive and non-responsive individuals.
Constructing Molecular Networks: Mapping Disease Mechanisms
Constructing molecular networks or pathways using multi-omics data allows researchers to infer interactions among genes, proteins, metabolites, and epigenetic marks. This provides a holistic view of disease mechanisms and drug actions, guiding the identification of potential therapeutic targets.
Prioritizing Drug Targets: Navigating the Sea of Biomolecules
The wealth of omics data aids in prioritizing potential drug targets based on various criteria such as differential expression, network centrality, functional annotation, disease, and drug associations. This step serves as a compass for researchers, steering them toward the most promising candidates for further exploration.
Validation through Experimentation: Bridging Bench to Bedside
Multi-omics not only identifies potential drug targets but also guides the validation process through experimental methods or computational models. Experiments such as knockdowns, overexpressions, mutations, inhibitors, activators, or combinations are designed based on multi-omics insights, serving as crucial steps in translating discoveries from the bench to the bedside.
Predicting Tomorrow: Multi-Omics in Drug Response Optimization
Beyond drug target identification, multi-omics plays a pivotal role in predicting and optimizing drug responses for diverse diseases. It characterizes inter-individual variability, classifies subtypes of individuals, and predicts optimal drug responses using machine learning methods.
Characterizing Inter-Individual Variability: The Genetic Tapestry of Drug Responses
Multi-omics identifies genetic variants, gene expression levels, protein expression levels, metabolite levels, and epigenetic modifications influencing inter-individual variability in drug responses. This knowledge provides a foundation for understanding how individuals respond to specific drugs.
Classifying Subtypes of Responders: Precision Medicine Unveiled
Clustering individuals based on molecular signatures of drug responses allows for the classification of responders versus non-responders, sensitive versus resistant, or toxic versus non-toxic groups. This step contributes to the burgeoning field of precision medicine, tailoring treatments to the individual.
Predicting Optimal Responses: Machine Learning as the Oracle
Multi-omics leverages machine learning methods like SVMs, random forests, or neural networks to build predictive models estimating the efficacy, safety, toxicity, adverse effects, resistance, sensitivity, dosage, and duration of drug responses. This heralds a future where treatments are optimized for each patient based on their unique molecular profile.
Triumphs in Multi-Omics: Illuminating the Path Ahead
Several pioneering studies underscore the transformative potential of multi-omics in elucidating complex biological phenomena. A study dissecting post-mortem brain samples elucidated the roles of risk-factor genes in diseases like autism spectrum disorder and Parkinson’s. Another study explored the intricate interactions between microbial metagenomes and host biology, shedding light on the influence of the microbiome on physiology, metabolism, immunity, and behavior.
In cancer research, multi-omics proved instrumental in predicting survival and drug response in breast cancer patients by integrating genomic, transcriptomic, proteomic, and epigenomic data from tumor-infiltrating immune cells. Similarly, a study delving into the molecular mechanisms of meningioma, a benign brain tumor, utilized multi-omics to unravel the functional roles of mutated genes, TRAF7 and KLF4, offering novel insights into disease understanding and therapeutic targeting.
As we navigate the vast landscape of biological intricacies, multi-omics emerges as a guiding light, promising not only to decipher the complexities of diseases and drug responses but also to shape the future of personalized medicine, innovative therapies, and the engineering of novel biological systems. In the realm of drug discovery, the integration of multi-omics stands poised as the key to unlocking.
Study DOI: 10.3390/proteomes11040034
Engr. Dex Marco Tiu Guibelondo, B.Sc. Pharm, R.Ph., B.Sc. CpE
Editor-in-Chief, PharmaFEATURES

Subscribe
to get our
LATEST NEWS
Related Posts

Bioinformatics & Multiomics
Mapping the Invisible Arrows: Unraveling Disease Causality Through Network Biology
What began as a methodological proposition—constructing causality through three structured networks—has evolved into a vision for the future of systems medicine.

Bioinformatics & Multiomics
Open-Source Bioinformatics: High-Resolution Analysis of Combinatorial Selection Dynamics
Combinatorial selection technologies are pivotal in molecular biology, facilitating biomolecule discovery through iterative enrichment and depletion.













